Dec 22, 2016
By Himanshu Joshi, MBBS, PhD
USC Norris Comprehensive Cancer Center, Keck School of Medicine
Novel molecular insights from cancer -omics data have proven that P4 (predictive, personalized, preventive, and participatory) precision medicine can be realized by the application of molecular diagnostics, therapeutics, prognostics, and prevention strategies tailored to alterations that predispose, drive, or propagate cancer. Molecular tumor boards (MTBs) are at the core of the institutional efforts of translating the -omics discoveries into precision medicine interventions. There are perhaps 20 to 30 active MTBs nationwide. In addition, several centers are operating MTBs in the formats of conventional tumor boards, virtual panel reviews, or clinical trials. Interest in application of precision medicine is growing, with discoveries of novel cancer-driving alterations and an expanding armamentarium of precision medicine alternatives. While the field is still in its infancy, genomic discoveries are emerging as a new dimension to seek solutions when conventional lines of therapy are not beneficial.
Which patients should be referred to the MTB? How should we interpret and select the driver growth circuitry and corresponding actionable targets out of a set of somatic and germ-line mutations, insertions and deletions, copy number alterations, and structural variations? What predicts clinical response corresponding to the therapeutic target? Having limited clinical evidence, many such questions appear to be a jigsaw puzzle. However, optimism is growing since the release of the Cancer Moonshot Initiative Blue Ribbon Panel recommendations, which are anticipated to foster coordination, collaboration, data sharing, and common standards of practice among institutional initiatives, such as the MTBs.
Current focus and clinical impact
MTBs mainly focus on cases that either involve complex decision-making or are less likely to respond (or are resistant) to conventional lines of therapy, and make clinical recommendations based on the -omic (mainly genomic or proteomic) profile(s) of cancers. MTBs may also help the care team optimize the diagnosis, determine therapeutic and preventive strategies tailored to germ-line predisposing events, assess patient eligibility for clinical trials, and predict prognosis.
One of the leading MTBs at University of California, San Diego (UCSD) Moores Cancer Center reported that 17 (40%) of the 43 reviewed patients received an MTB-recommended line of therapy. Of these 17 patients, 41% experienced stable disease of at least 6 months or partial remission.1 Further, a meta-analysis of 570 phase II single-agent clinical trials in multiple cancer types (representing approximately 32,000 patients) comparing personalized versus non-personalized arms showed a higher median response rate (31% vs. 10.5%, respectively; p < 0.001), prolonged median progression-free survival (5.9 vs. 2.7 months, respectively; p < 0.001) and overall survival (13.7 v. 8.9 months, respectively; p < 0.001).2 Additional studies can provide a better idea on what types/profiles of cancers can benefit the most from targeted therapy alternatives.
The University of North Carolina Lineberger Comprehensive Cancer Center is another example of a large collaborative effort that has sequenced more than 10,000 cancer samples, including 2,000 samples for patient care and approximately 8,000 research subjects contributing to the Cancer Genome Atlas. David N. Hayes, MD, MPH, director of clinical bioinformatics at Lineberger Cancer Center, said that in a small but measurable percentage of cancers, often poorly differentiated, genomics could find molecular patterns indicative of a different tumor than what was described by the routine pathology reports. Examples include poorly differentiated carcinomas being redefined as melanoma, or clarification of cancer type in the setting of a patient with multiple primary cancers. He noted that sequencing has proven helpful in making subtle but important diagnostic distinctions, such as differentiating adenocarcinoma of the lung from squamous cell carcinoma, or uterine cancer from an endometrioid variant of ovarian cancer, or detection of HPV in a tumor that was clinically found HPV-negative.
Challenges
A variety of medical, technologic, logistical, regulatory, legal, financial, ethical, and social issues have been limiting the scope of targeted therapeutics.
Smart cancers often present with aggressive mutator phenotypes and genomic instability arising from alterations in caretaker (tumor suppressors) or landscaper (microenvironment) genes coinciding with overlapping or linked alterations in growth signaling, rendering the tumors resistant to conventional modes of therapy. While molecular review may help optimize the clinical management, these tumors are highly adaptive in acquiring alternative growth signaling, thus making sustained response a challenge.
Questions of targeting what, when, and why pose a key challenge because of the small number of clinically actionable alterations and the many unknowns due to lack of sufficient data on patient outcomes, safety, toxicity, and pharmacogenomic profile. According to John D. Carpten, PhD, chair of the USC Translational Genomics Institute, greater coordination and collaboration with the pharmaceutical industry is essential for evaluating the potential benefit that novel drugs can offer in the context of precision oncology trials.
Distinction between the whole exome/genome versus specialized multigene panels must be made in view of genes and type of alterations of interest on a case-to-case basis, with a goal of maximizing the sensitivity and specificity. Without adequate power, the detection of rare variation with low allele fraction could be a challenge. Sequencing the matched germ line, and relevant companion diagnostic and validation assays with demonstrated clinical validity (when available), may be of help.
Clinical manifestations as well as response may vary considerably, even when a similar driver gene is involved, for reasons such as mutation position and type and epigenomic factors. Therefore, bringing the individual-level precision in the current scenario may appear unrealistic.
Off-label use of targeted agents for similar alterations in another tumor type/tissue has not been proven beneficial; there are concerns about safety and serious toxicities.3 The same applies to tissue-specific feedback mechanisms. Vemurafenib is effective in BRAF (V600E) melanoma, but not in colorectal cancer because of the EGFR activation.4
Implementation of follow-up tumor profiling in case of any new event may be challenging for a variety of reasons, including the cost of profiling.
Prioritization of targets in case of more than one actionable alteration is a common issue, particularly because of the lack of evidence on anticipated patient outcome by comparing alternative targets. Follow-up tumor profiling of relapse and recurrence is necessary to identify the target(s) that predict superior outcome and lower risk of relapse/recurrence. With an estimated 5% annual recurrence rate 5 to 10 years after diagnosis and thereafter maintaining a stable rate of slightly less than 3%,5 hormone receptor–positive breast cancers pose a question of identifying the predictors and drivers of late recurrence.
Cancer genomes consist of multiple competing clones that acquire novel mutations for acquiring growth and survival advantage. A single biopsy may not be sufficient to represent subclonal drivers that can switch to an alternative growth signaling pathway.
Some of the practical issues include a lapse of time between the profiling and review, variations in available expertise, and lack of universal adaption to a common reporting standard (i.e., the American College of Medical Genetics [ACMG]6) and a common reporting format.
Computation pipelines and variant classification practices may vary considerably in absence of consensus benchmarking criteria. Use of peer-reviewed open-source approaches and compliance with common guidelines (i.e., ACMG6) can promote inter-institution comparability of the results.
Collective wisdom and the Cancer Moonshot
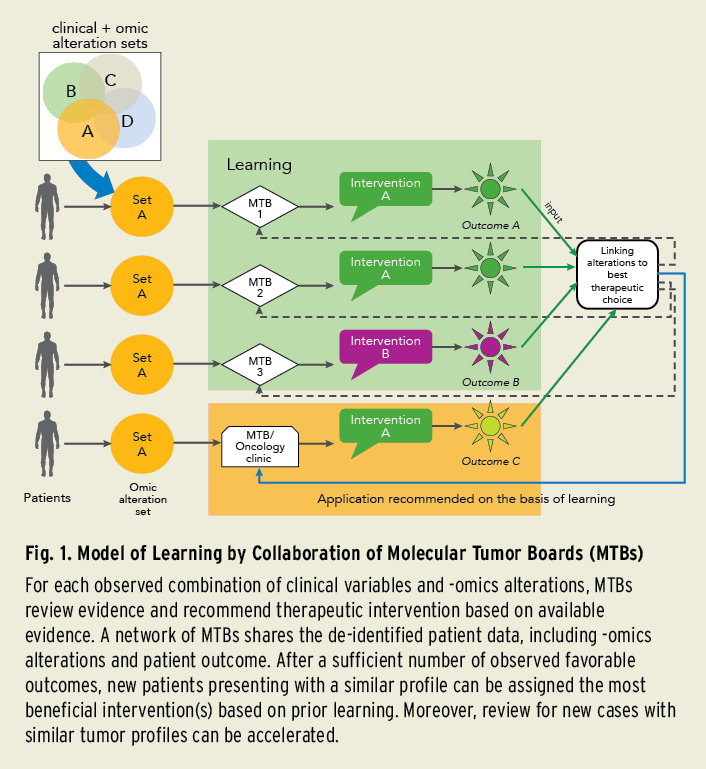 Building up the evidence is a priority. The Cancer Moonshot Initiative Blue Ribbon Panel recommendations emphasize the creation of a national data ecosystem and 3D cancer atlas for enabling improved data sharing among researchers and clinicians, as well as promoting data mining. Projects such as the NCI Genomic Data Commons and ASCO’s CancerLinQ® are steps in this direction. MTBs, by sharing the expert-reviewed best practice recommendations together with the patient data, can accelerate collective learning (Fig. 1).
Building up the evidence is a priority. The Cancer Moonshot Initiative Blue Ribbon Panel recommendations emphasize the creation of a national data ecosystem and 3D cancer atlas for enabling improved data sharing among researchers and clinicians, as well as promoting data mining. Projects such as the NCI Genomic Data Commons and ASCO’s CancerLinQ® are steps in this direction. MTBs, by sharing the expert-reviewed best practice recommendations together with the patient data, can accelerate collective learning (Fig. 1).
When following matching treatment guidelines, disparities in care by volume of cancer centers can make a 10% to 15% difference in patient outcome.7,8 Strategies such as dose optimization and toxicity management may considerably affect the patient outcome. In addition to benefitting from the shared expertise on precision medicine, small-volume centers can benefit by sharing expertise on various other aspects of cancer care through networking and data sharing.
John L. Marshall, MD, chair of the CARIS Centers of Excellence for Precision Medicine Network™ (which specializes in collaborative efforts for tumor profiling and recommending the best possible intervention), is looking forward to more public-private partnerships as a path towards success in precision medicine.
Equally important is knowledgeable national leadership addressing scientific and legal aspects of genomic testing and targeted therapies, said Razelle Kurzrock, MD, who leads the personalized medicine efforts at UCSD. Dr. Kurzrock also noted a need to create an umbrella protocol that permits proper patient consent for data sharing.
Promoting the development of disease-specific panels for deep sequencing the genomic regions of interest is necessary to enhance the validity of evidence and thereby empower MTBs. Dr. Hayes views a need for regulatory guidance from the U.S. Food and Drug Administration and the U.S. Patent and Trademark Office for accelerating the development and implementation of new diagnostic technologies.
Conclusion
As the national leadership and guidance improves, networking the MTBs can prove to be an important step to accelerate our mission of changing the landscape of cancer care.
References
- Parker BA, Schwaederlé M, Scur MD, et al. J Oncol Pract. 2015;11:442-9.
- Schwaederle M, Zhao M, Lee JJ, et al. J Clin Oncol. 2015;33:3817-25.
- Levêque D. Lancet Oncol. 2008;9:1102-7.
- Prahallad A, Sun C, Huang S, et al. Nature. 2012;483:100-3.
- Colleoni M, Sun Z, Price KN, et al. J Clin Oncol. 2016;34:927-35.
- Richards S, Aziz N, Bale S, et al. Genet Med. 2015;17:405-24.
- Eaton BR, Pugh SL, Bradley JD, et al. J Natl Cancer Inst. 2016;108.


